Researchers at MIT have developed a cutting-edge computational model that enhances the ability of large language models (LLMs) to predict antibody structures with remarkable accuracy. This innovation could revolutionize the development of antibody-based drugs, targeting a wide array of infectious diseases, including SARS-CoV-2.
The Challenge of Antibody Prediction
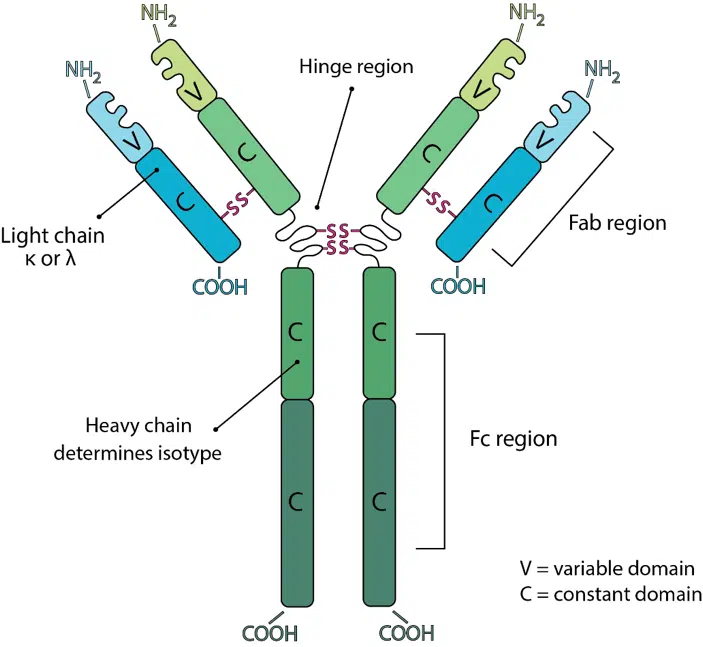
While LLMs such as AlphaFold have significantly improved the prediction of general protein structures, antibodies present unique challenges. Their hypervariable regions—critical for detecting and binding to antigens—are highly diverse and not constrained by evolutionary patterns. This variability complicates structure prediction, as these regions often escape the predictive power of traditional AI models.
To address this gap, MIT researchers created a new technique that focuses specifically on these hypervariable regions, paving the way for more accurate structural predictions and better-targeted therapies.
The AbMap Model
The researchers’ new computational model, named AbMap, leverages advanced LLM techniques and incorporates two key modules:
- Sequence Similarity Module
Trained on hypervariable sequences from 3,000 antibody structures, this module identifies patterns that correlate with structural similarities. - Binding Strength Module
This module correlates 3,700 antibody sequences with their binding strength to specific antigens, enabling predictions of how effectively antibodies can neutralize targets like the SARS-CoV-2 spike protein.
By integrating these modules, AbMap not only predicts antibody structures but also evaluates their binding efficiency, offering a dual advantage for researchers.
Real-World Impact
Using AbMap, researchers generated millions of antibody variants by modifying hypervariable regions, identifying those with the highest potential for binding success. Experimental tests confirmed that 82% of the antibodies predicted by AbMap outperformed the original candidates, demonstrating the model’s utility in drug discovery.
This approach has significant implications for pharmaceutical development. By identifying promising candidates early, companies can avoid costly failures in preclinical trials and develop a diverse portfolio of options to mitigate risks during drug development.
Beyond Drug Discovery
The applications of this model extend beyond therapeutic development. AbMap can also help researchers explore immune responses across individuals, shedding light on why some people respond differently to infections like HIV or Covid-19. By analyzing the structural repertoire of antibodies, researchers can gain deeper insights into immune system dynamics and uncover strategies for improving vaccine efficacy.
A Scalable and Accurate Solution
AbMap bridges the gap between sequence-based and structure-based analysis, offering a scalable tool for understanding antibody behavior. This breakthrough aligns with the broader goals of using AI to address critical challenges in healthcare, making it a promising tool for advancing precision medicine.
Sources: https://news.mit.edu/2025/new-computational-model-can-predict-antibody-structures-more-accurately-0102, https://bioxcell.com/educational-articles/antibody-structure/